Giuseppe Tosto, MD, a neurologist and a genetic epidemiologist at NewYork-Presbyterian/
Late-onset Alzheimer’s disease (LOAD), in which symptoms typically become apparent after age 65, is the most common cause of neurodegenerative disease worldwide, but it is not equally distributed among racial and ethnic groups. The frequency of the disease in African Americans and Caribbean Hispanics is significantly higher than in non-Hispanic Whites. While the ε4 isoform of apolipoprotein E (APOE) gene increases a person’s risk of developing the disease, it alone does not explain the ethnic disparities of LOAD. Ancestry, a critical aspect of health disparities, may help to explain the observed differences in frequency of LOAD across ethnic groups.
“Understanding the genetic risk and protective factors contributing to Alzheimer’s disease will help clarify which of these factors are specific to one ethnic group or are universally shared, and will ultimately help us develop drugs for prevention and treatment that are better tailored to the individual and are therefore much more effective,” explains Dr. Tosto, assistant professor of Neurology in the Taub Institute for Research on Alzheimer’s Disease and the Aging Brain at NewYork-Presbyterian/
The researchers analyzed data from three studies that recruited individuals of Caribbean Hispanic (CH) ancestry: (1) the Washington Heights and Inwood Columbia Aging Project (WHICAP study); (2) Estudio Familiar de Influencia Genetica en Alzheimer (EFIGA); and (3) the Puerto Rico 10/66 study (10/66 PR). The large cohort of CH (N=8,813) were from New York, the Dominican Republic, and Puerto Rico. The researchers used admixture mapping, a method used to prioritize ancestral blocks associated with diseases in ethnic groups from two or more geographically distinct ancestral populations. Admixture mapping is particularly useful in this scenario because it can be used to compare differences in local ancestry at a specified genetic locus when performing association with a disease, in contrast to another type of genetic analyses, genome-wide association studies (GWAS), which measure differences in allele frequencies between cases and controls. The analyses were conducted separately for ancestral components (European [EUR], African [AFR], Native American[NA]) and jointly (AFR+NA).
Figure 1. Manhattan plots for the Joint Admixture Mapping analyses highlighting the genome-wide significant ancestral blocks.
After identifying the significant ancestral blocks, the blocks were fine-mapped using multi-ethnic whole-exome sequencing data (“WES,” a type of genetic sequencing) from WHICAP to identify rare variants associated with LOAD and replicated in the UK Biobank WES dataset. The candidate genes were validated studying protein expression in human LOAD and control brains, and in two animal Alzheimer’s disease models, Drosophila and Zebrafish.
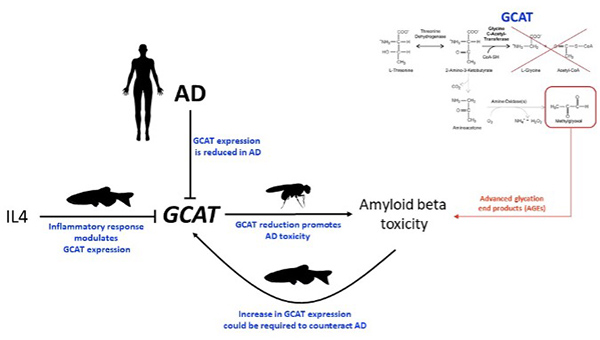
Admixture mapping uncovered several genomic regions harboring new LOAD-associated loci that the researchers say might explain the observed different frequency of LOAD across ethnic groups. In the joint AFR+NA model, the researchers identified four significant ancestral blocks located on chromosomes 1 (p-value=8.94E-05), 6 (p-value=8.63E-05), 21 (p-value=4.64E-05) and 22 (p-value=1.77E-05). Fine-mapping using WHICAP prioritized the GCAT gene on chromosome 22 (SKAT-O p-value=3.45E-05) and replicated in the UK Biobank (SKAT-O p-value=0.05). In LOAD brains, a decrease of 28% in GCAT protein expression was observed (p-value=0.038), and GCAT knockdown in Amyloid-β42 Drosophila exacerbated rough eye phenotype (an indicator of AD) (68% increase, p-value=4.84E-09). In zebrafish, gcat expression increased after acute amyloidosis (34%, p-value=0.0049), and decreased upon anti-inflammatory Interleukin-4 (39%, p-value=2.3E-05). These data propose GCAT activity as an amyloid-induced beneficial response, which cannot be sustained in humans.
Figure 2. GCAT deficiency in Alzheimer’s disease pathogenesis. The figure shows integration of human ex-vivo analysis with functional experiments conducted in Drosophila and Zebrafish brains.
This study underscores the power of admixture mapping and subsequent fine mapping for uncovering genetic regions harboring new LOAD-associated loci, which may explain the observed differences in frequency of LOAD across ethnic groups. In the era of personalized medicine, this discovery will help to address inequalities and medical disparities in LOAD treatment and prevention.