Maximizing Molecular Coding
NCI-MATCH Trial
NewYork-Presbyterian participates in the National Cancer Institute-based MATCH trial and is represented on the program’s Executive Committee. A nationwide effort to sequence 7,000 patients with different types of cancer across the country, MATCH is a precision medicine cancer treatment clinical trial in which patients are assigned to receive treatment based on the genetic changes found in their tumors through genomic sequencing and other tests. Now near completion, the MATCH trial has identified 30 different drugs correlated to the different genes of patients, making it possible for patients anywhere in the country to have access to treatment specific to the genetic makeup of their tumor.
The NCI-COG Pediatric MATCH launched in July 2017 with two of the eight treatment arms of the program led by Columbia physicians.
Building on Mutation-Based Therapy

Dr. Andrea Califano
With the keen awareness that mutation-based therapy does not yet have a widespread application to cancer care, the need for a more comprehensive strategy in cancer treatment has emerged throughout the field. Researchers at Columbia are now exploring RNA-based, systems biology approaches that significantly complement the strength of therapeutic programs and collaborations at Columbia based both on mutational analysis – with its focus on whole-genome DNA sequencing – and on immuno-oncology.
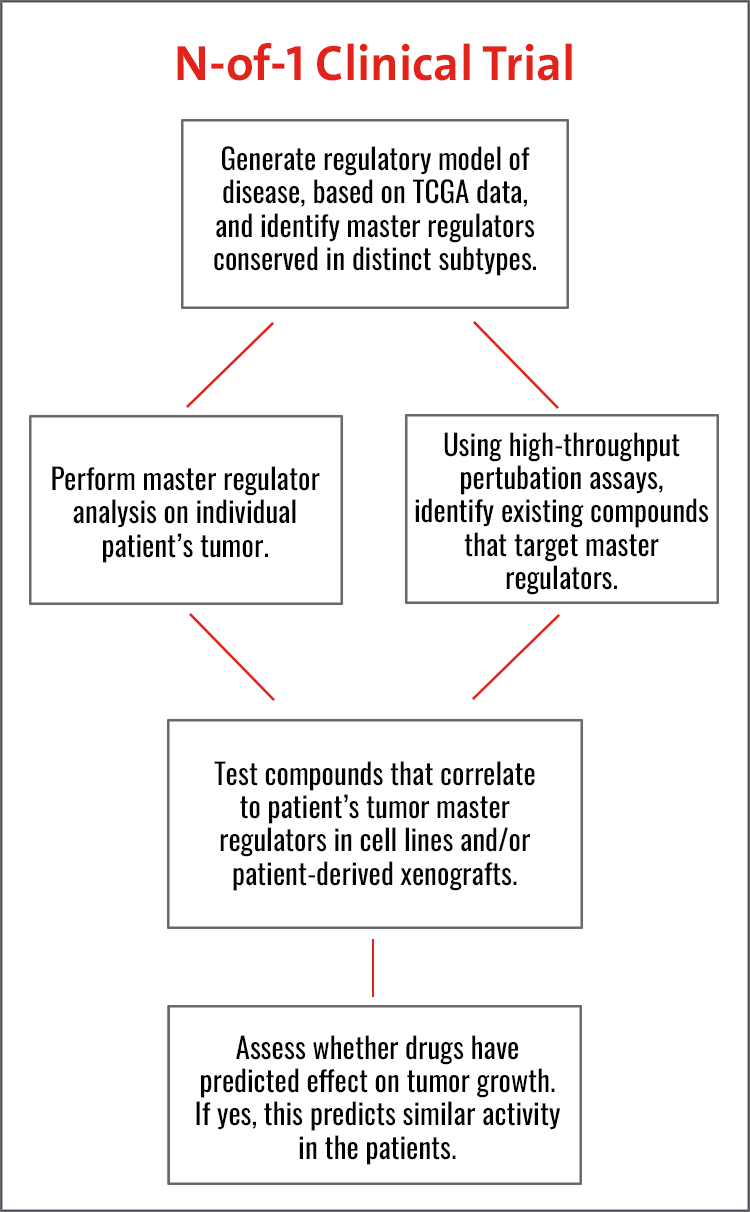
Their work builds on the increasing success of their N-of-1 trial program in which FDA-approved and other clinically relevant investigational compounds are being prioritized based on their effectiveness in blocking master regulator proteins responsible for tumor state maintenance. Conducted in individual patients, who are progressing after multiple lines of standard-of-care therapy, these clinical trials have been exploratory in nature to determine the specific genetic and molecular factors that are essential for a cancer’s growth in a single patient.
N-of-1 trials currently underway or in development at Columbia target 14 adult human malignancies, from metastatic prostate, ovarian and breast cancers to glioblastoma and anaplastic meningioma, and are being extended to pediatric tumors, starting with sarcomas. Critically, the two technologies they developed to support these studies, OncoTarget and OncoTreat, recently received New York State Department of Health CLIA certification and are available for clinical studies through the Columbia Department of Pathology and Cell Biology.

To address a particular hurdle in these trials in which patients are referred at an advanced stage of their disease, generally following failure of several other therapies, the researchers are now collaborating with clinicians specializing in ovarian and breast cancer to use the algorithm in the neoadjuvant phase when the tumor is still very vulnerable. Another challenge is getting access to specific drugs prioritized by the study. To address this issue, the researchers are now looking at the N-of-1 study as a potential pre-selection phase for enrollment into a second, more traditional clinical trial where the patient will be able to receive the specific drug.
At this point in time, the Columbia researchers note that there are only 24 master regulator modules (dubbed tumor checkpoints) that cover key dependencies of about 20,000 patients whose samples have been collected by The Cancer Genome Atlas (TCGA), for which RNA and DNA profiles are already available. Buoyed by this information, an enormous amount of progress has been made in the clinic. For example, in the N-of-1 trials 34 drugs were predicted by two algorithms developed at Columbia – OncoTarget™ and OncoTreat™ – for patients who had failed from three to seven lines of therapy and were considered terminal. When the drugs were tested in direct mouse transplants of the human tumor, 20 of the 34 drugs induced an objective response, resulting in either stable or reduced tumor volume. Of four patients who could receive one of the drugs predicted by the analysis, three had a clinical response.
OncoTarget
This core, New York State Department of Health CLIA certified test systematically assesses whether any of more than 100 proteins for which a targeted drug already exists are aberrantly activated in a patient-specific tumor sample independent of the tumor’s DNA mutational state.
OncoTreat
This core, New York State Department of Health CLIA certified test systematically prioritizes FDA-approved drugs and investigational agents for a patient-specific tumor. This is accomplished by aligning their tumor-specific activity against the full repertoire of master regulator proteins that represent the “engine room” (tumor checkpoint) of a patient-specific tumor. OncoTreat is currently being deployed in clinical trials at leading cancer research centers.
Application to Pediatrics

The pediatric oncology program at NewYork-Presbyterian/Columbia is one of the front-runners nationally instituting and implementing real-time precision oncology for patients, including the establishment in 2014 of a precision in pediatric sequencing program called the PIPseq Cancer Program. Through PIPseq, every child with a high-risk cancer that comes to Columbia undergoes whole-exome sequencing; the patient’s tumor also undergoes RNA-seq analysis. In 2016, the Columbia program received funding from the Sohn Conference Foundation to provide this clinical testing to any child with a high-risk cancer in the New York, New Jersey, Connecticut area.
The comprehensive platform includes an assessment of the genes that code for proteins in both a patient’s tumor and normal tissue. Tumor-specific genomic alterations can be identified, providing clinicians with information on cancer variance, cancer-specific mutations, copy number variations, translocations and fusions, and gene expression data, as well as a window into inherited cancer predisposition should families choose to learn this information. In 2018, the program will start integrating the OncoTarget and OncoTreat methodologies originally developed for the adult N-of-1 program. Columbia is among only a handful of institutions nationwide providing this type of platform for pediatric patients.
As of spring 2017, 250 patients have been tested. Potential targets were found in about 40 percent of patients; information of diagnostic significance in about 15 percent of patients; and a germline predisposition in about 14 percent of patients. Overall, the testing revealed something that was clinically impactful in about two of every three patients.
Under Columbia’s Developmental Therapeutics Program for Pediatrics, 20 to 30 phase 1 and 2 clinical trials are enrolling patients at any one time to provide access to the drugs identified as relevant.
Evolution of Urothelial Carcinoma

Dr. Bishoy M. Faltas
Researchers at Weill Cornell have identified how selective pressure from chemotherapy directs the evolution of urothelial carcinoma and shapes its clonal architecture – a key biological question with clinical implications. Their whole-exome sequencing analysis of the clonality of 72 urothelial carcinoma samples, including 16 matched sets of primary and advanced tumors that were prospectively collected before and after chemotherapy, provided the following insights:
-
chemotherapy-treated urothelial carcinoma is characterized by intra-patient mutational heterogeneity, and the majority of mutations are not shared
-
branching evolution and metastatic spread are very early events in the natural history of urothelial carcinoma
-
chemotherapy-treated urothelial carcinoma is enriched with clonal mutations involving the L1-cell adhesion molecule and integrin signaling pathways
-
APOBEC-induced mutagenesis is clonally enriched in chemotherapy-treated urothelial carcinoma and continues to shape the evolution of urothelial carcinoma throughout its lifetime

Columbia Combined Cancer Panel
The Columbia Combined Cancer Panel (CCCP), which is approved by the New York State Department of Health, is one of the few such clinical oncology panels in the New York area. A sequencing-only test, the CCCP was designed in collaboration with Columbia oncologists and is conducted within Columbia University Irving Medical Center’s Personalized Genomic Medicine CLIA-certified clinical genomics laboratory.
This panel queries 467 cancer-related genes and is a critical assay for our mission to deliver the highest quality of cancer care to our patients. By routinely profiling the tumors of patients with advanced malignancies, we more completely understand the unique biology of each tumor and have the ability to tailor care appropriately. Through our Experimental Therapeutics Clinic, we have access to approved and investigational targeted therapies that would be predicted to be effective for our patients based upon the molecular profile of their disease, and thus we are now truly able to deliver precision oncologic care to our patients.
Single-Cell Omics

The CCCP uses the Illumina HiSeq2500 platform, a sequencing-only test, to query 467 cancer-related genes.
At Weill Cornell, researchers have integrated genome sequencing into the care of cancer patients. They are now pursuing a better understanding of cancer through the behavior of single cells and integrating single cell transcriptomics in their precision medicine workflow. Their work involves leveraging new technologies to analyze clinical samples and visualize various diseased tissues at single-cell resolution and to isolate and sequence the genes that are expressed within individual cells across thousands of tumor and non-tumor cells.
By dissecting the complexity of the immune cells and different types of immune cell populations within tumors and determining how they interact with tumor cells, the researchers can rapidly diagnose patients and predict which treatments might work. These include treatments that may use the immune system to kill cancer cells, which treatments may encounter resistance, and how the disease is likely to progress.
The researchers are also developing new precision medicine technologies that examine DNA and cells from a blood draw or urine sample from one patient, predict his or her future risk for developing a specific type of cancer, or reveal presence of a hidden tumor.


